freealg Documentation#
freealg is a Python package that employs free probability to evaluate the spectral densities of large matrix forms. The fundamental algorithm employed by freealg is free decompression, which extrapolates from the empirical spectral densities of small submatrices to infer the eigenspectrum of extremely large matrices.
Install#
Install with pip:
pip install freealg
Alternatively, clone the source code and install with
cd source_dir
pip install .
Quick Usage#
Generate a matrix with its spectral density following Marcheno-Pastur law
using freealg.distributions.MarchenkoPastur:
>>> from freealg import FreeForm
>>> from freealg.distributions import MarchenkoPastur
>>> import numpy
>>> mp = MarchenkoPastur(1/50)
>>> A = mp.matrix(3000)
>>> eig = numpy.linalg.eigvalsh(A)
Create a freealg.FreeForm object
>>> from freealg import FreeForm
>>> ff = FreeForm(eig, support=(mp.lam_m, mp.lam_p))
Fit its density using Chebyshev method using freealg.FreeForm.fit().
Alternative method is Jacobi polynomial.
>>> psi = ff.fit(method='chebyshev', K=10, alpha=2, beta=2, reg=0,
... damp='jackson', force=True, pade_p=1, pade_q=1, plot=True)
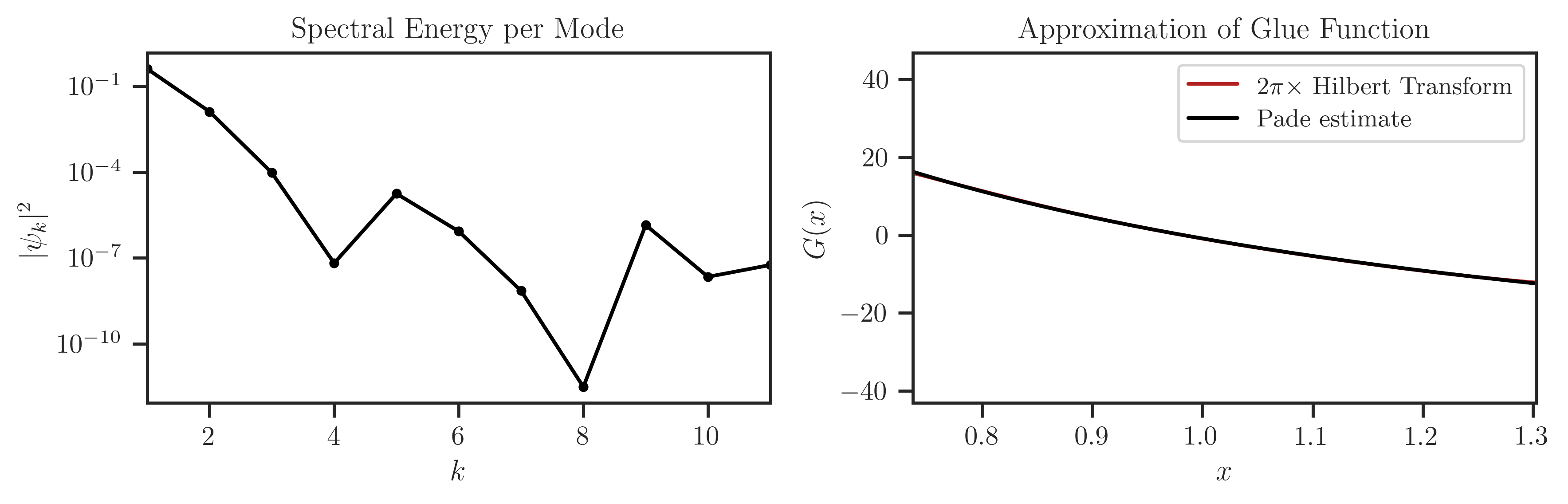
Estimate density using freealg.FreeForm.density:
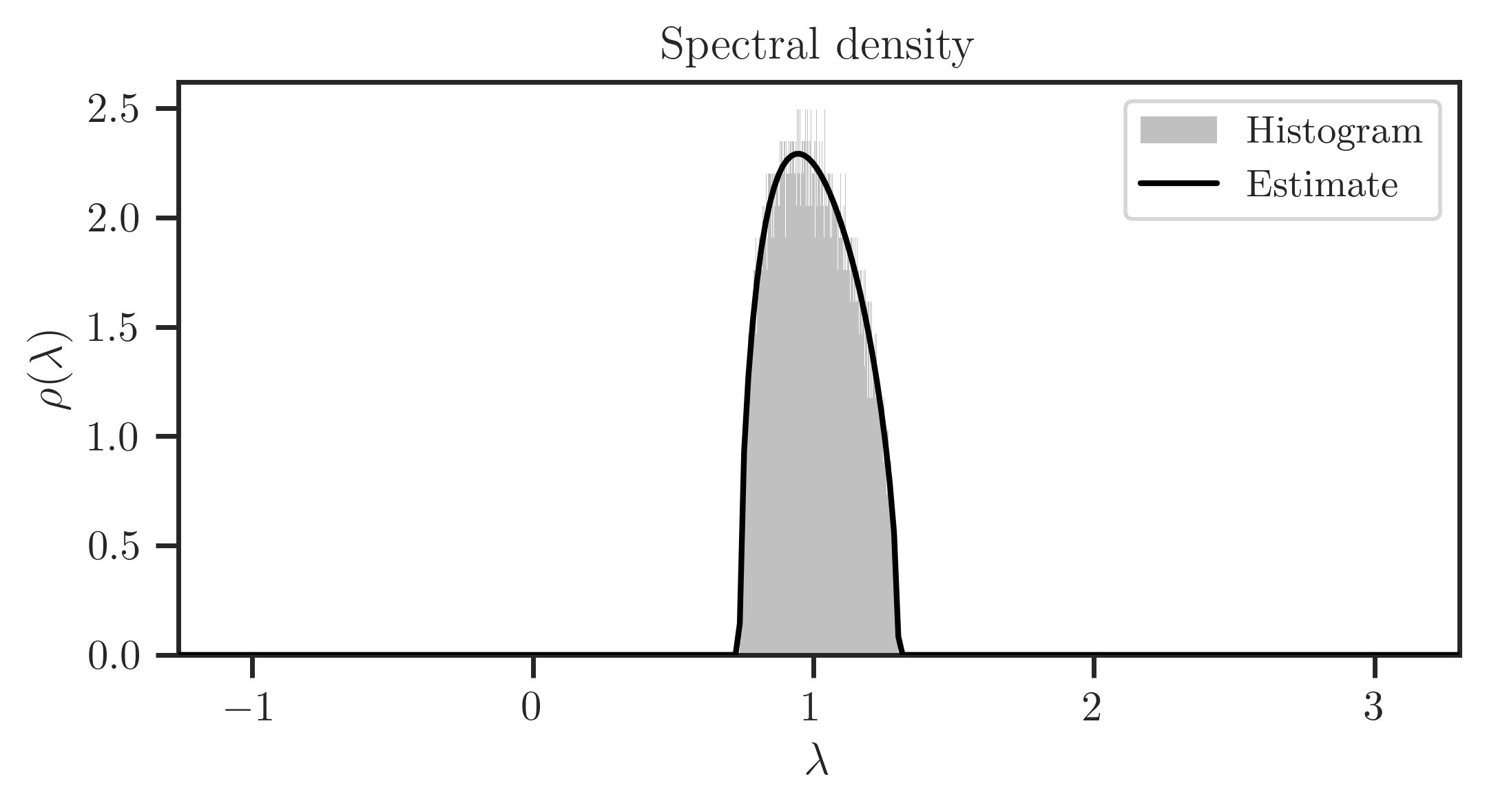
>>> x = numpy.linspace(lam_m-2, lam_p+2, 300)
>>> rho = ff.density(x, plot=True)
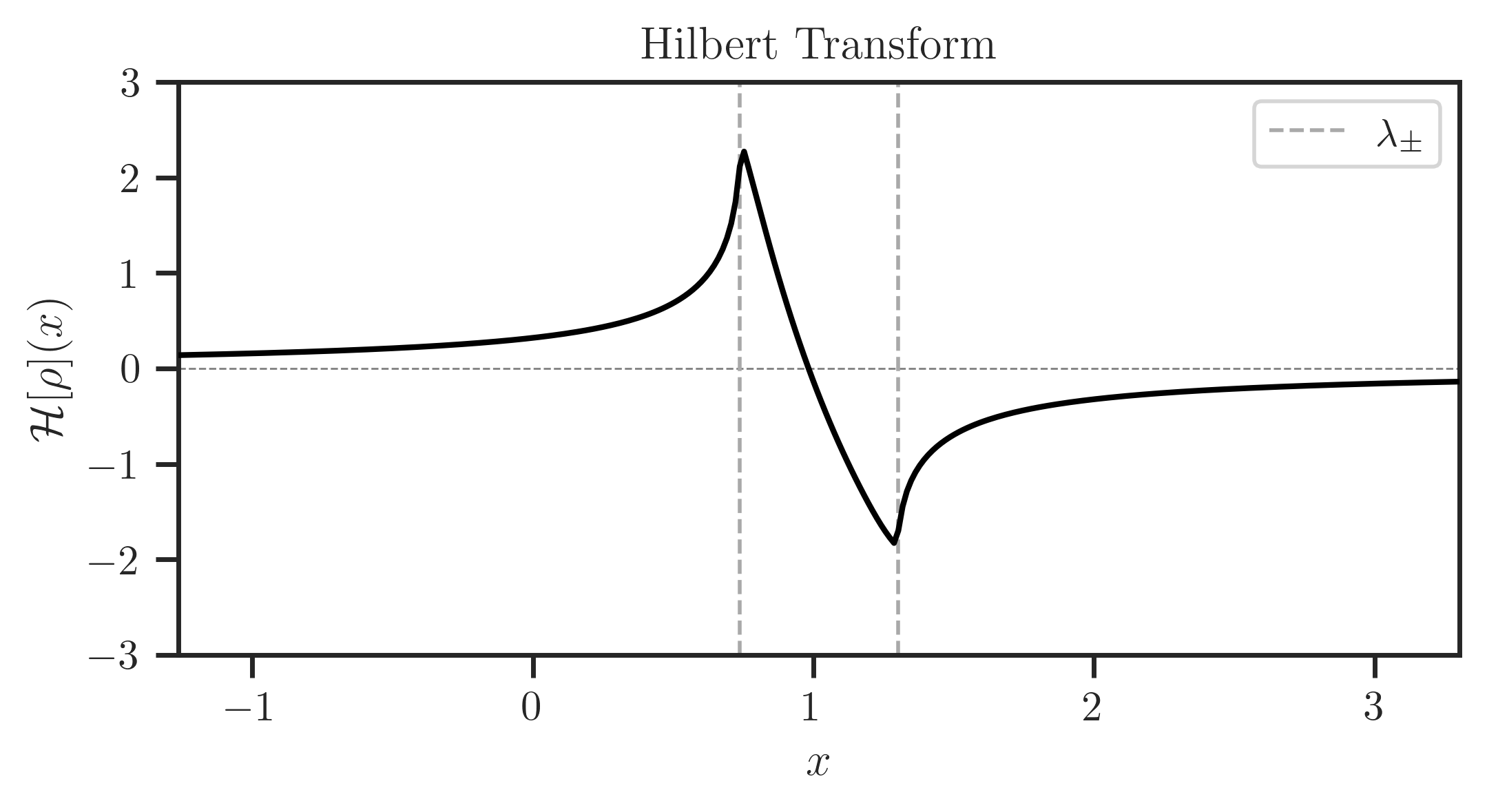
Estimate Hilbert transform using freealg.FreeForm.hilbert:
>>> hilb = ff.hilbert(x, rho, plot=True)
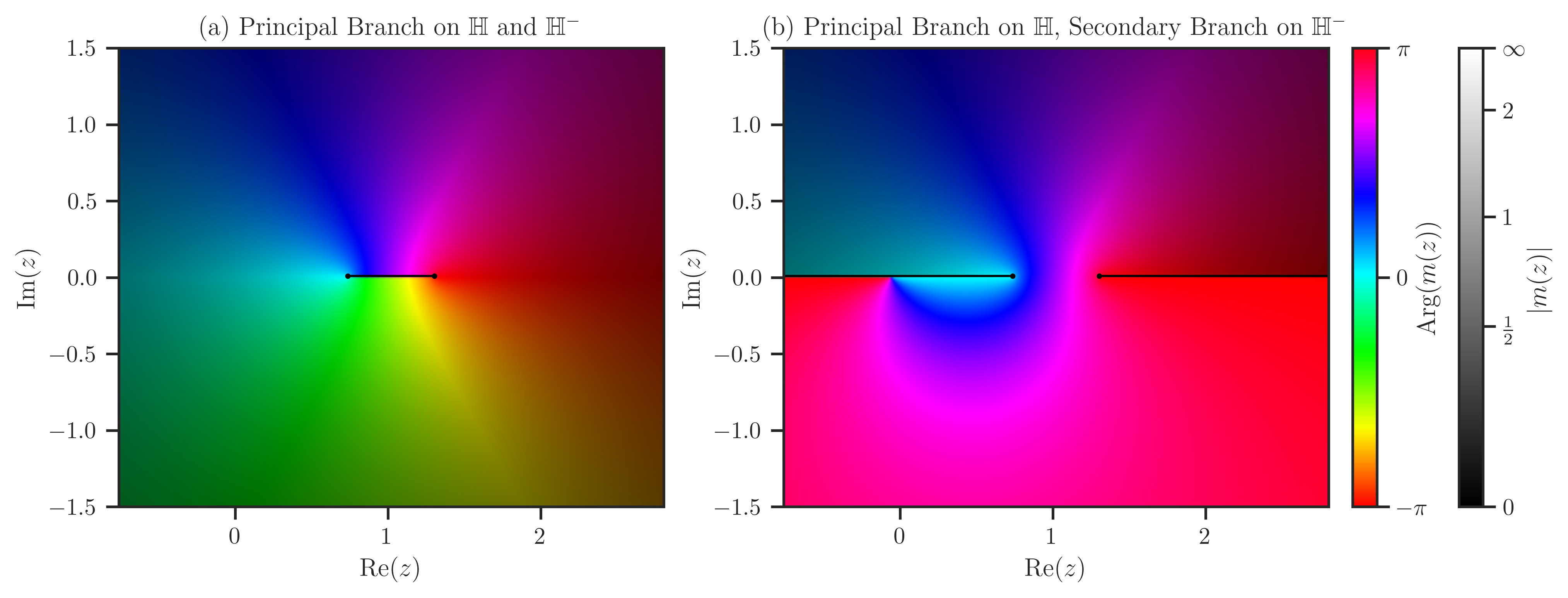
Estimate Stieltjes transform with freealg.FreeForm.stieltjes:
>>> x = numpy.linspace(lam_m-1.5, lam_p+1.5, 300)
>>> y = numpy.linspace(-1.5, 1.5, 200)
>>> mp, mm = ff.stieltjes(x, y, plot=True)
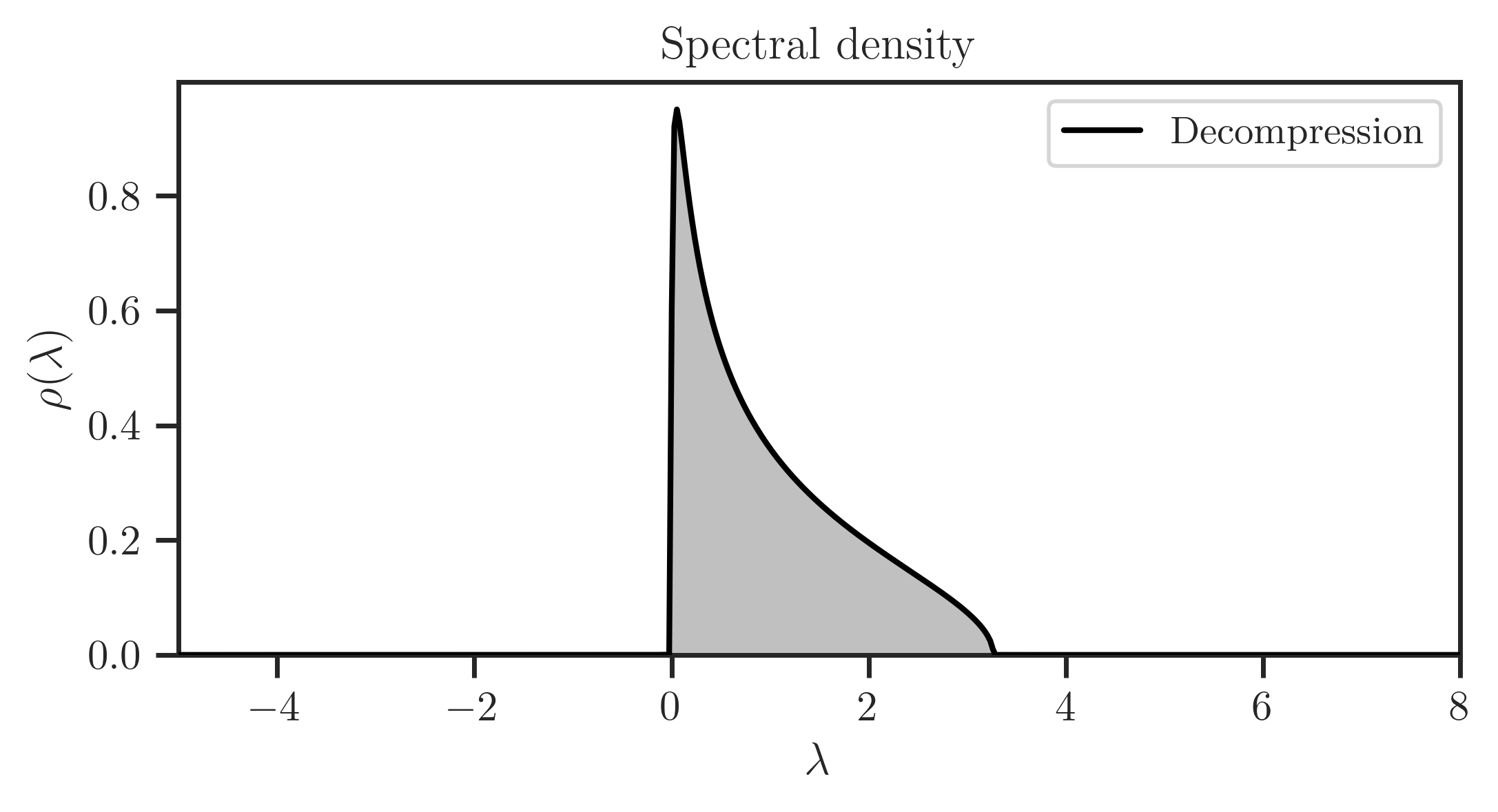
Decompress spectral density to a larger matrix with
freealg.FreeForm.decompress():
>>> rho, x = ff.decompress(100_000, plot=True)
API Reference#
Check the list of functions, classes, and modules of freealg with their usage, options, and examples.
Test#
You may test the package with tox:
cd source_dir
tox
Alternatively, test with pytest:
cd source_dir
pytest
How to Contribute#
We welcome contributions via GitHub’s pull request. Developers should review our Contributing Guidelines before submitting their code. If you do not feel comfortable modifying the code, we also welcome feature requests and bug reports.


