glearn.priors.Uniform#
- class glearn.priors.Uniform(a=0, b=inf)#
Uniform distribution.
Note
For the methods of this class, see the base class
glearn.priors.Prior.- Parameters:
- afloat or array_like[float], default=0
The left point of an interval \([a, b]\) of the uniform distribution. If
ais given as an array \((a_1, \dots, a_p)\), the prior is assumed to be \(p\) independent distributions, each on the interval \([a_i, b_i]\).- bfloat or array_like[float], default=numpy.inf
The right point of an interval \([a, b]\) of the uniform distribution. If
bis given as an array \((b_1, \dots, b_p)\), the prior is assumed to be \(p\) independent distributions, each on the interval \([a_i, b_i]\).
See also
Notes
Single Hyperparameter:
The uniform distribution in the interval \([a, b]\) is defined by the probability density function
\[\begin{split}p(\theta) = \begin{cases} 1, & a \leq \theta \leq b, \\ 0, & \text{otherwise}. \end{cases}\end{split}\]Multiple Hyperparameters:
If an array of the hyperparameters are given, namely \(\boldsymbol{\theta} = (\theta_1, \dots, \theta_p)\), then the prior is the product of independent priors
\[p(\boldsymbol{\theta}) = p(\theta_1) \dots p(\theta_p).\]In this case, if the input arguments
aandbare given as the arrays \(\boldsymbol{a} = (a_1, \dots, a_p)\) and \(\boldsymbol{b} = (b_1, \dots, b_p)\), each prior \(p(\theta_i)\) is defined as the uniform distribution on the interval \([a_i, b_i]\). In contrary, ifaandbare given as the scalars \(a\) and \(b\), then all priors \(p(\theta_i)\) are defined as uniform distributions in the interval \([a, b]\).Examples
Create Prior Objects:
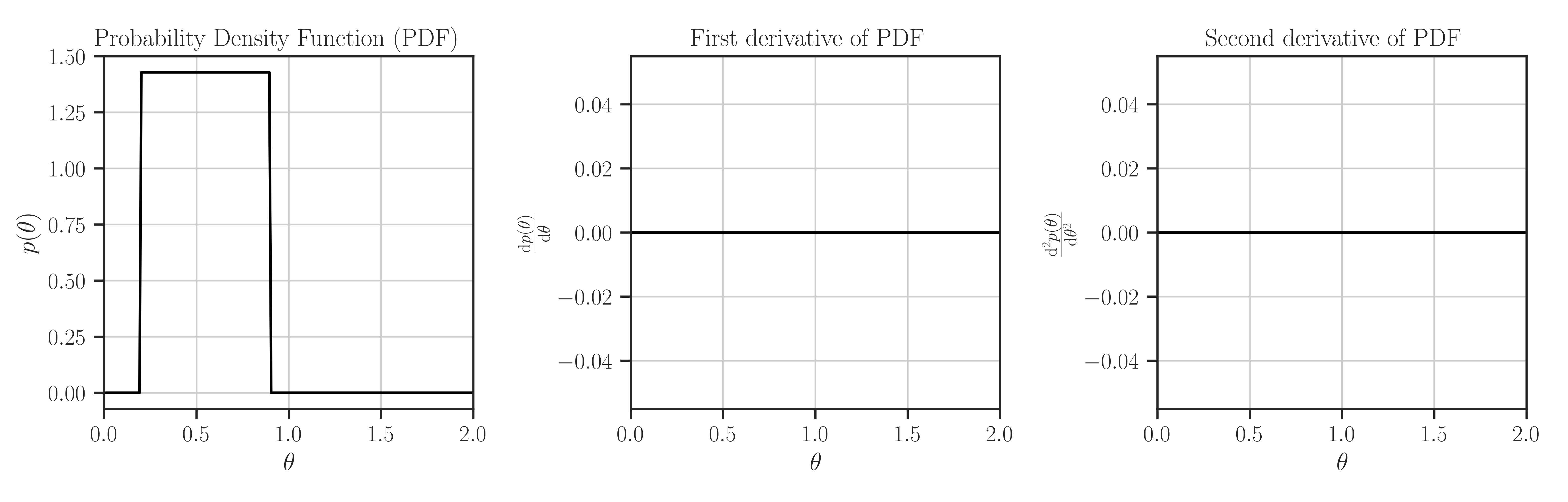
Create uniform prior in the interval \([0.2, 0.9]\):
>>> from glearn import priors >>> prior = priors.Uniform(0.2, 0.9) >>> # Evaluate PDF function at multiple locations >>> t = [0, 0.5, 1] >>> prior.pdf(t) array([0. , 1.42857143, 0. ]) >>> # Evaluate the Jacobian of the PDF >>> prior.pdf_jacobian(t) array([0., 0., 0.]) >>> # Evaluate the Hessian of the PDF >>> prior.pdf_hessian(t) array([[0., 0., 0.], [0., 0., 0.], [0., 0., 0.]]) >>> # Evaluate the log-PDF >>> prior.log_pdf(t) -inf >>> # Evaluate the Jacobian of the log-PDF >>> prior.log_pdf_jacobian(t) array([nan, nan, nan]) >>> # Evaluate the Hessian of the log-PDF >>> prior.log_pdf_hessian(t) array([[nan, 0., 0.], [ 0., nan, 0.], [ 0., 0., nan]]) >>> # Plot the distribution and its first and second derivative >>> prior.plot()
Where to Use the Prior object:
Define a covariance model (see
glearn.Covariance) where its scale parameter is a prior function.>>> # Generate a set of sample points >>> from glearn.sample_data import generate_points >>> points = generate_points(num_points=50) >>> # Create covariance object of the points with the above kernel >>> from glearn import covariance >>> cov = glearn.Covariance(points, kernel=kernel, scale=prior)
- Attributes:
- afloat or array_like[float], default=0
Input argument a
- bfloat or array_like[float], default=0
Input argument b
- meanfloat or array_like[float], default=0
Mean of the distribution
Methods
suggest_hyperparam([positive])Find an initial guess for the hyperparameters based on the peaks of the prior distribution.
pdf(x)Probability density function of the prior distribution.
pdf_jacobian(x)Jacobian of the probability density function of the prior distribution.
pdf_hessian(x)Hessian of the probability density function of the prior distribution.