imate.InterpolateTrace#
- class imate.InterpolateTrace(A, B=None, p=-1, options={}, verbose=False, kind='imbf', ti=[], **kwargs)#
Interpolate the trace of the powers of an affine matrix function.
- Parameters:
- Anumpy.ndarray, scipy.sparse matrix
Symmetric positive-definite matrix (positive-definite if p is non-positive). Matrix can be dense or sparse.
Warning
Symmetry and positive (semi-) definiteness of A will not be checked. Make sure A satisfies these conditions.
- Bnumpy.ndarray, scipy.sparse matrix, default=None
Symmetric positive-definite matrix (positive-definite if p is non-positive). Matrix can be dense or sparse. B should have the same size and type of A. If B is None (default value), it is assumed that B is the identity matrix.
Warning
Symmetry and positive (semi-) definiteness of B will not be checked. Make sure B satisfies these conditions.
- pfloat, default=-1
The power \(p\) of the matrix which can be real positive or negative, but not zero. For zero power see
imate.InterpolateLogdet.- optionsdict, default={}
At each interpolation point \(t_i\), the trace is computed using the following functions:
imate.trace()(if \(p>0\))imate.traceinv()(if \(p < 0\)).
The
optionspasses a dictionary of arguments to the above functions.- verbosebool, default=False
If True, it prints some information about the computation process.
- kind{‘ext’, ‘eig’, ‘mbf’, ‘imbf’, ‘rbf’, ‘crf’, ‘spl’, ‘rpf’}, default: ‘imbf’
The algorithm of interpolation, which are the same as the algorithms in
imate.InterpolateSchatten. See documentation for each specific algorithm:- tifloat or array_like(float), default=None
Interpolation points, which can be a single point, a list, or an array of points. The interpolator honors the exact function values at the interpolant points. If an empty list is given, i.e.,
[], depending on the algorithm, a default list of interpolant points is used. Also, the size of the array of ti depends on the algorithm as follows. Ifkindis:extoreig, the notiis required.mbf, then a single pointtimay be specified.imbf,spl, orrbf, then a list oftiwith arbitrary size may be specified.crforrpf, then a list oftipoints with even size may be specified.
- kwargs**kwargs
Additional arguments to pass to each specific algorithm. See documentation for each
kindin the above.
Notes
Interpolation of Affine Matrix Function:
This class interpolates the trace of powers of the one-parameter matrix function:
\[t \mapsto \left( \mathbf{A} + t \mathbf{B} \right)^p,\]where the matrices \(\mathbf{A}\) and \(\mathbf{B}\) are symmetric and positive semi-definite (positive-definite if \(p < 0\)) and \(t \in [t_{\inf}, \infty)\) is a real parameter where \(t_{\inf}\) is the minimum \(t\) such that \(\mathbf{A} + t_{\inf} \mathbf{B}\) remains positive-definite.
The interpolator is initialized by providing \(q\) interpolant points \(t_i\), \(i = 1, \dots, q\), which are often given in a logarithmically spaced interval \(t_i \in [t_1, t_p]\). The interpolator can interpolate the above function at arbitrary inquiry points \(t \in [t_1, t_p]\) using various methods.
References
[1]Ameli, S., and Shadden. S. C. (2022). Interpolating Log-Determinant and Trace of the Powers of Matrix \(\mathbf{A} + t \mathbf{B}\). Statistics and Computing 32, 108. https://doi.org/10.1007/s11222-022-10173-4.
Examples
Basic Usage:
Interpolate the trace of the affine matrix function \((\mathbf{A} + t \mathbf{B})^{-1}\) using
imbfalgorithm and the interpolating points \(t_i = [10^{-2}, 10^{-1}, 1, 10]\).>>> # Generate two sample matrices (symmetric and positive-definite) >>> from imate.sample_matrices import correlation_matrix >>> A = correlation_matrix(size=20, scale=1e-1) >>> B = correlation_matrix(size=20, scale=2e-2) >>> # Initialize interpolator object >>> from imate import InterpolateTrace >>> ti = [1e-2, 1e-1, 1, 1e1] >>> f = InterpolateTrace(A, B, p=-1, kind='imbf', ti=ti) >>> # Interpolate at an inquiry point t = 0.4 >>> t = 4e-1 >>> f(t) 20.56849471404019
Alternatively, call
imate.InterpolateTrace.interpolate()to interpolate at points t:>>> # This is the same as f(t) >>> f.interpolate(t) 20.56849471404019
To evaluate the exact value of the trace at point t without interpolation, call
imate.InterpolateTrace.eval()function:>>> # This evaluates the function value at t exactly (no interpolation) >>> f.eval(t) 20.625083538250383
It can be seen that the relative error of interpolation compared to the exact solution in the above is \(0.27 \%\) using only four interpolation points \(t_i\), which is a remarkable result.
Warning
Calling
imate.InterpolateTrace.eval()may take a longer time to compute as it computes the function exactly. Particularly, if t is a large array, it may take a very long time to return the exact values.Arguments Specific to Algorithms:
In the above example, the
imbfalgorithm is used. See more arguments of this algorithm at imate.InterpolateSchatten(kind=’imbf’). In the next example, we passbasis_func_typespecific to this algorithm:>>> # Passing kwatgs specific to imbf algorithm >>> f = InterpolateTrace(A, B, p=-1, kind='imbf', ti=ti, ... basis_func_type='ortho2') >>> f(t) 20.56849471404019
You may choose other algorithms using
kindargument. For instance, the next example uses the Chebyshev rational functions with an additional argumentfunc_typespecific to this method. See more details at imate.InterpolateSchatten(kind=’crf’).>>> # Generate two sample matrices (symmetric and positive-definite) >>> f = InterpolateTrace(A, B, p=-1, kind='crf', ti=ti, func_type=1) >>> f(t) 20.63092837950084
Passing Options:
The above examples, the internal computation is passed to
imate.traceinv()function since \(p=-1\) is negative. You can pass arguments to the latter function usingoptionsargument. To do so, create a dictionary with the keys as the name of the argument. For instance, to use imate.trace(method=’slq’) method withmin_num_samples=20andmax_num_samples=100, create the following dictionary:>>> # Specify arguments as a dictionary >>> options = { ... 'method': 'slq', ... 'min_num_samples': 20, ... 'max_num_samples': 100 ... } >>> # Pass the options to the interpolator >>> f = InterpolateTrace(A, B, p=-1, options=options, kind='imbf', ... ti=ti) >>> f(t) 20.43067873913403
You may get a different result than the above as the slq method is a randomized method.
Interpolate on Range of Points:
Once the interpolation object
fin the above example is instantiated, callingimate.InterpolateTrace.interpolate()on a list of inquiry points t has almost no computational cost. The next example inquires interpolation on 1000 points t:Interpolate an array of inquiry points
t_array:>>> # Create an interpolator object again >>> ti = [1e-2, 1e-1, 1, 1e1] >>> f = InterpolateTrace(A, B, p=-1, ti=ti) >>> # Interpolate at an array of points >>> import numpy >>> t_array = numpy.logspace(-2, 1, 1000) >>> norm_array = f.interpolate(t_array)
One may plot the above interpolated results as follows:
>>> import matplotlib.pyplot as plt >>> # Plot settings (optional) >>> from imate._utilities import set_custom_theme >>> set_custom_theme(font_scale=1.15) >>> plt.semilogx(t_array, norm_array, color='black') >>> plt.xlim([t_array[0], t_array[-1]]) >>> plt.ylim([0, 40]) >>> plt.xlabel('$t$') >>> plt.ylabel('$\mathrm{trace} (\mathbf{A} + t \mathbf{B})^{-1}$') >>> plt.title('Interpolation of Trace of Inverse') >>> plt.show()

Plotting Interpolation and Compare with Exact Solution:
A more convenient way to plot the interpolation result is to call
imate.InterpolateTrace.plot()function.>>> f.plot(t_array)
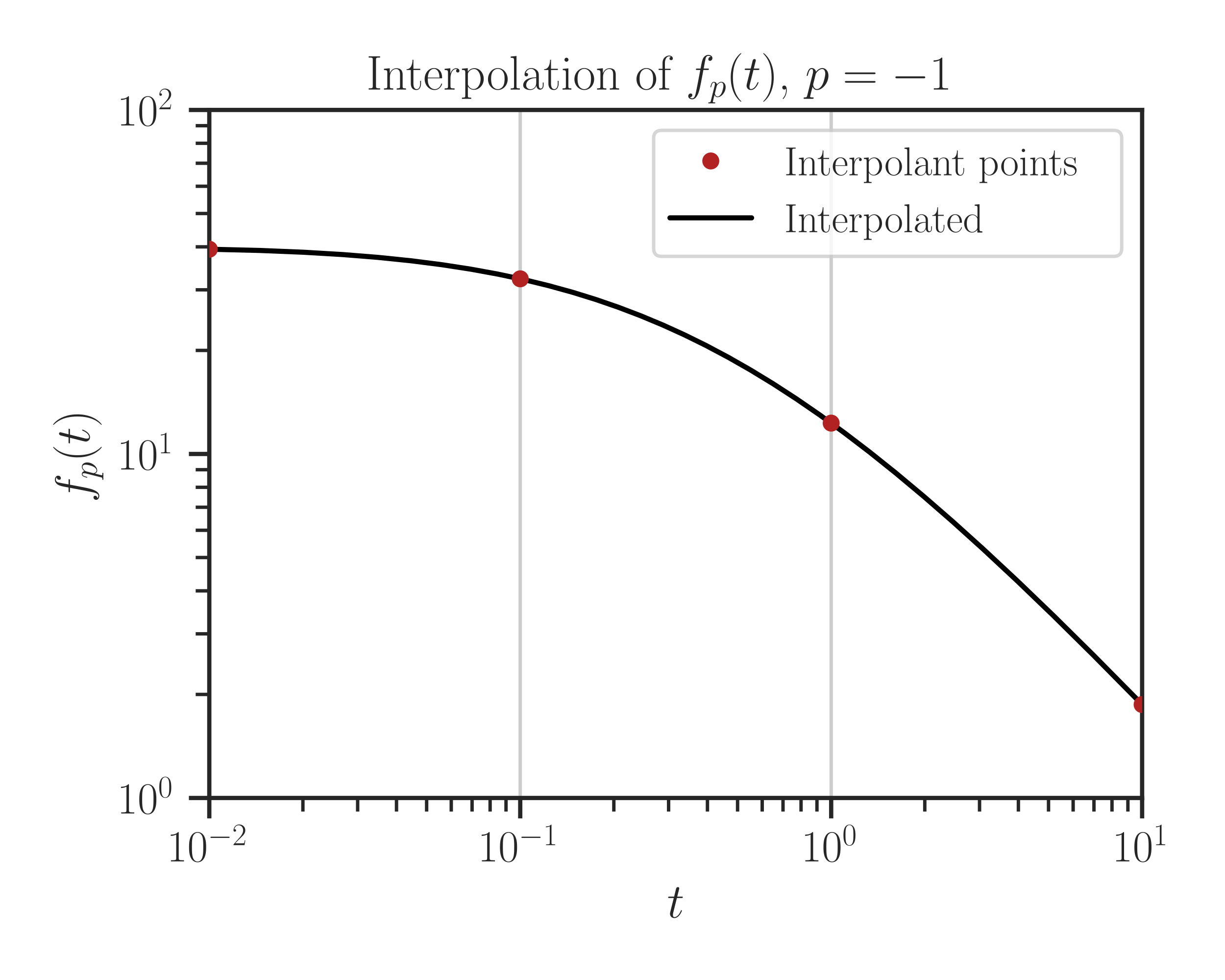
In the above, \(f_p(t) = \mathrm{trace} (\mathbf{A} + t \mathbf{B})^p\) is the Schatten norm. If you set
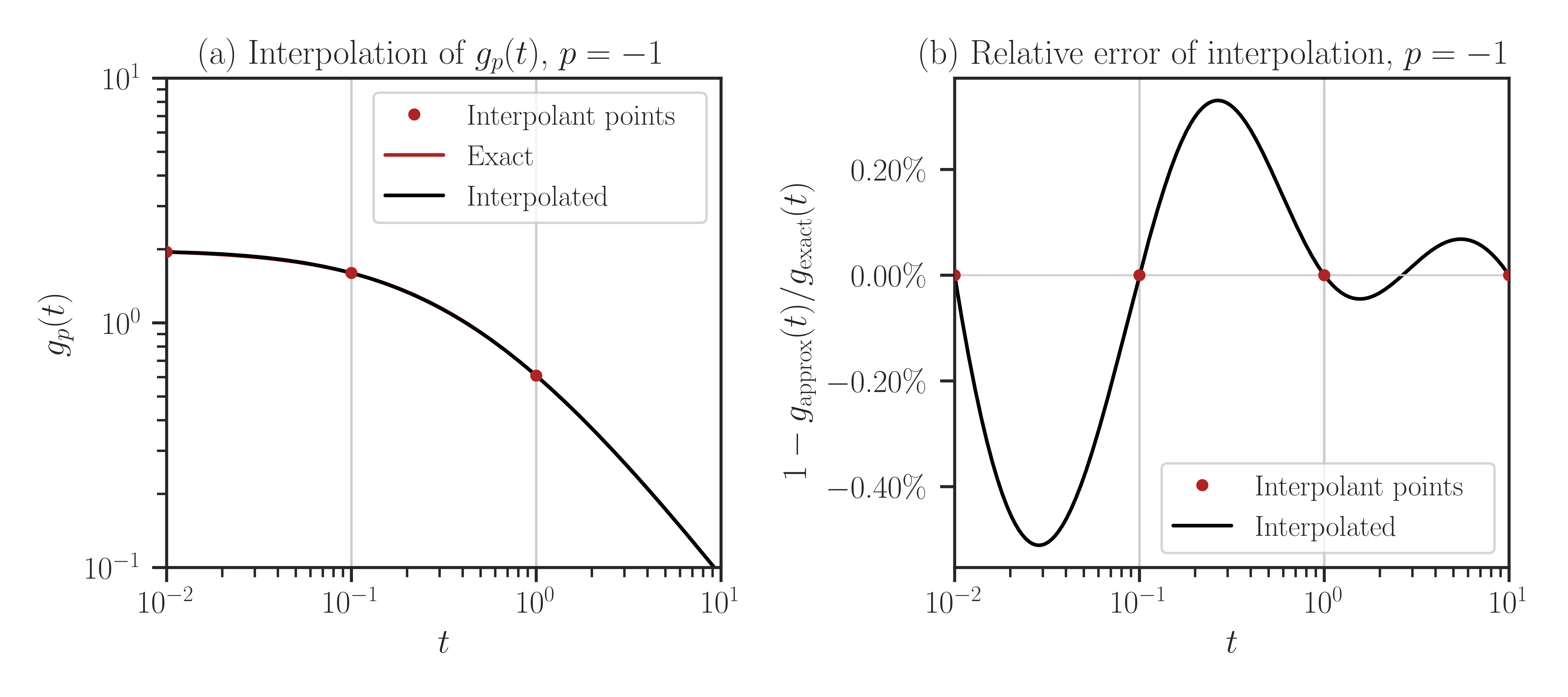
normalizeto True, it plots the normalized function\[g_p(t) = \frac{\mathrm{trace}(\mathbf{A} + t \mathbf{B})^{p}}{ \mathrm{trace}(\mathbf{B})^p}.\]To compare with the true values (without interpolation), pass
compare=Trueto the above function.Warning
By setting
compareto True, every point in the array t is evaluated both using interpolation and with the exact method (no interpolation). If the size of t is large, this may take a very long run time.>>> f.plot(t_array, normalize=True, compare=True)
- Attributes:
- kindstr
Method of interpolation
- verbosebool
Verbosity of the computation process
- nint
Size of the matrix
- qint
Number of interpolant points
- pfloat
The power \(p\) of the matrix.
Methods
__call__(t)Interpolate at the input point t.
eval(t)Evaluate the exact value of the function at the input point t without interpolation.
interpolate(t)Interpolate at the input point t.
Returns the scale parameter of the interpolator.
upper_bound(t)Upper bound of the interpolation function.
lower_bound(t)Upper bound of the interpolation function.
plot(t[, normalize, compare])Plot the interpolation results.